Methodology
Genome Assembly and Annotation Pipeline
The Citrus reticulata var. Murcott genome was assembled at the chromosomal level through a comprehensive bioinformatics pipeline integrating PacBio HiFi long-reads, Illumina short-reads, and Hi-C data. The workflow comprised stringent quality control, read filtering and taxonomic classification, primary assembly with HiFiasm, assembly purification to eliminate organellar contaminants, and Hi-C–based scaffolding to achieve chromosome-scale resolution.
Gene prediction and annotation followed a multi-phase strategy, including repetitive element masking with RepeatModeler2 and RepeatMasker, ab initio gene prediction with the Helixer deep learning framework, protein domain characterization using InterProScan5, signal peptide detection with Phobius, and extensive functional annotation through Funannotate, with Gene Ontology (GO) term assignment and UniProt cross-referencing.
Assembly quality was assessed with BUSCO and QUAST, yielding a high-quality reference genome with 98.2% BUSCO completeness. This work represents a concerted effort by the Chilean biotechnology company Meristem, aimed at enhancing the identification of genes associated with agronomically relevant traits, thereby providing a solid foundation for their subsequent editing and the development of new, improved citrus varieties.
Assembly Statistics
| Metric | Value | Description |
|---|---|---|
| Total Assembly Size | 322.8 Mbp | Total length of assembled sequences |
| Number of Contigs | 9 | Count of contiguous assembled sequences |
| Contig N50 | 38.1 Mbp | Length above which 50% of the assembly is contained in contigs |
| Longest Contig | 49.7 Mbp | Length of the largest assembled contig |
| GC Content | 36.4% | Proportion of guanine and cytosine bases in the genome |
| BUSCO Completeness | 98.2% | Percentage of complete orthologs recovered |
| Predicted Genes | 27,043 | Number of predicted protein-coding genes |
| Repeat Content | 53.65% | Proportion of the genome composed of repetitive elements |
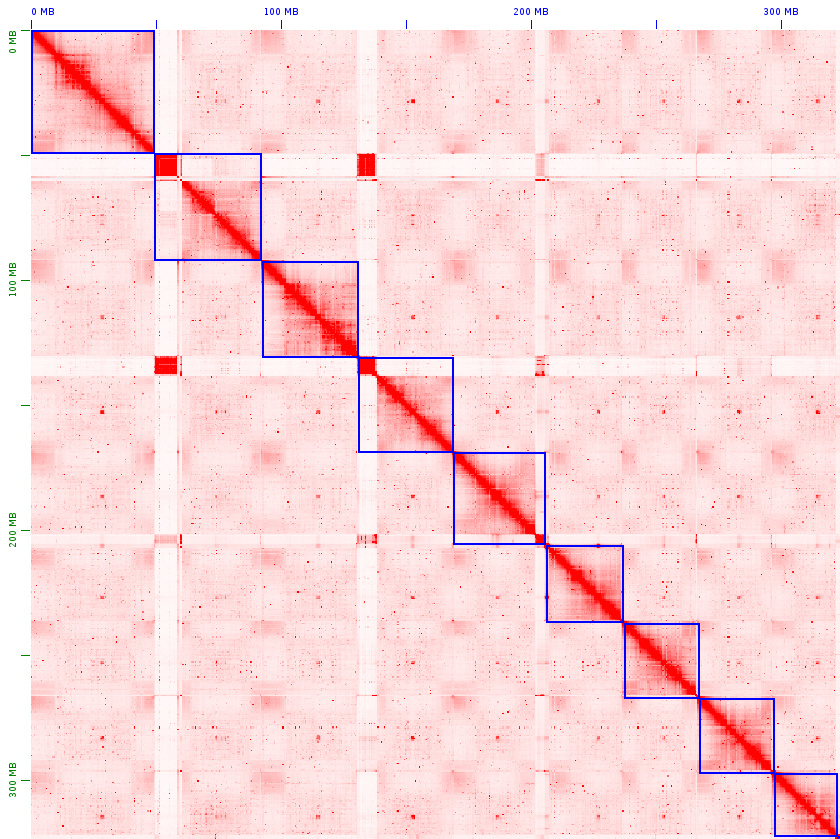
Hi-C Contact Maps of the 9 Chromosomes
These heatmaps display the chromatin contact frequencies across the nine chromosomes, derived from Hi-C data. The intensity of each region reflects the level of interaction, allowing the visualization of genomic architecture and large-scale structural organization.
Genome Browser
Explore the Citrus reticulata var. Murcott genome using the interactive genome browser below. Navigate across chromosomes, zoom into specific regions, and examine diverse genomic features such as genes, annotations, and variants.
